Pipelines¶
The overall goal of MLBlocks is to be able to build Pipelines.
What is a Pipeline?¶
A Pipeline is a sequence of primitives working together to learn from training data and later on make predictions on new data as if they were a single object.
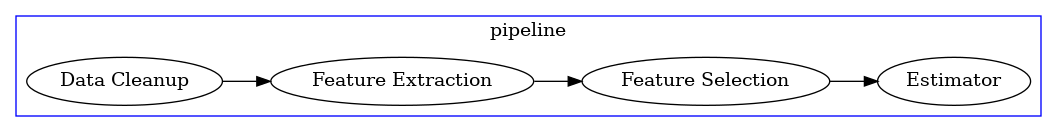
Pipelines can have any possible composition, but the typical setup includes data cleanup, feature extraction and feature selection primitives, and ends up with one or more estimator primitives that generate the final predictions.
Also, just like primitives, pipelines have hyperparameters, which are nothing but the list of hyperparameters that the primitives that compose them expect, and which can be also tuned to improve their prediction performance.
The MLPipeline Class¶
In MLBlocks, a pipeline is represented by the mlblocks.MLPipeline class, which combines
multiple mlblocks.MLBlock instances, called blocks in this context, and calls them in
succession for fitting and predicting.
As you have seen in the quickstart tutorial, You can create an MLPipeline by simply passing it the list of primitives that will compose it:
In [1]: from mlblocks import MLPipeline
In [2]: primitives = [
...: 'sklearn.preprocessing.StandardScaler',
...: 'sklearn.ensemble.RandomForestClassifier'
...: ]
...:
In [3]: pipeline = MLPipeline(primitives)
Block Names¶
When the MLPipeline is created, a list of MLBlock instances has been created inside it.
Each of this blocks is given a unique name which is composed by the primitive name and a counter,
separated by a hash symbol #, allowing multiple blocks for the same primitive to exist
within a single MLPipeline:
In [4]: dict(pipeline.blocks)
Out[4]:
{'sklearn.preprocessing.StandardScaler#1': <mlblocks.mlblock.MLBlock at 0x7f01e0a81160>,
'sklearn.ensemble.RandomForestClassifier#1': <mlblocks.mlblock.MLBlock at 0x7f01e08c3f40>}
Init Arguments¶
Sometimes, additional arguments need to be passed to the blocks during initialization.
This can be done by passing an extra dictionary to the MLPipeline when it is created:
In [5]: init_params = {
...: 'sklearn.preprocessing.StandardScaler': {
...: 'with_mean': False
...: },
...: 'sklearn.ensemble.RandomForestClassifier': {
...: 'n_estimators': 100
...: }
...: }
...:
In [6]: pipeline = MLPipeline(primitives, init_params=init_params)
This dictionary must have as keys the name of the blocks that the arguments belong to, and as values the dictionary that contains the argument names and their values.
Note
If only one block of a specific primitive exists in the pipeline, the counter appended to its name can be skipped when specifying the arguments, as shown in the example.
Context¶
One element that plays an important role during the execution of the fit and predict methods of a pipeline is the Context dictionary.
Each time any of these methods is called, a context dictionary is internally created and all the variables passed to the method are stored in it.
Then, the following happens for each block:
The list of arguments that the method expects is retrieved from the block configuration.
The corresponding values are read from the Context and passed to the method.
The list of outputs that the method returns is retrieved from the block configuration.
The indicated outputs are captured in order and put back to the Context dictionary using the name specified.
Context Usage Examples¶
The most simple version of this schema is one where all the blocks expect a single feature matrix as input, called X, and output another feature matrix, also called X, until the last one, which outputs the final prediction, called y.
In this case, supposing that we only have 3 blocks, the sequence when the pipeline.predict(X)
call is issued would be:
The value of
Xis stored in the Context.The value of
Xis pulled from the Context and passed to block1.The output from block1 is put back into the Context, overwriting the old value.
The value of
Xis pulled again from the Context and passed to block2.The output from block2 is put back into the Context, overwriting again the old value.
The value of
Xis pulled for the last time from the Context and passed to block3.The output from block3, since it is the last one, is returned.
![digraph G {
subgraph cluster_0 {
label = "pipeline.predict(X)";
b1 [label="block1.produce(X)"];
b2 [label="block2.produce(X)"];
b3 [label="block3.produce(X)"];
b1 -> b2 -> b3 [style=invis];
subgraph cluster_1 {
X1 [label=X];
X2 [label=X];
X3 [label=X];
X1 -> X2 -> X3 [style="dashed"];
label = "Context";
}
}
X -> X1;
X1 -> b1 [constraint=false];
b1 -> X2;
X2 -> b2 [constraint=false];
b2 -> X3;
X3 -> b3 [constraint=false];
b3 -> y;
}](../_images/graphviz-b3981e4480a82cc9f77d01d9337416ff795e60e5.png)
Another schema with some more complexity would be one where there is one primitive that needs to be passed an additional argument that provides information about the data.
Suppose, for example, that there is a primitive that encodes categorical features, but
it needs to be given the list of features that it needs to encode in a variable called
features. Suppose also that this primitive is followed directly by an estimator primitive.
In this case, the call would be pipeline.predict(X, features=features), and the sequence
of actions would be:
The value of
Xandfeaturesis stored in the Context.The value of
Xandfeaturesis pulled from the Context and passed to the encoder block.The output from encoder is put back into the Context as
X, overwriting the old value.The value of
Xis pulled again from the Context and passed to the estimator block.The output from the estimator block is returned.
![digraph G {
subgraph cluster_0 {
label = "pipeline.predict(X, features=features)";
b1 [label="encoder.produce(X, features=features)"];
b2 [label="estimator.produce(X)"];
b1 -> b2 [style=invis];
subgraph cluster_1 {
{rank=same X1 f1}
X1 [label=X group=c];
f1 [label=features group=c];
X2 [label=X group=c];
f1 -> X1 [style=invis];
X1 -> X2 [style=dashed];
label = "Context";
}
}
{rank=same X features}
features -> f1;
X -> X1;
X1 -> b1 [constraint=false];
f1 -> b1 [constraint=false];
b1 -> X2;
X2 -> b2 [constraint=false]
b2 -> y
}](../_images/graphviz-30b772283c09a682c0839163ff176f99cc84d262.png)
But, what if we also have a primitive, which we will call detector, that detects which features are categorical and want to use it instead of passing a manually crafted list of features?
We can also achieve it using the Context!
In this case, we go back to the pipeline.predict(X) call, and let the detector primitive
do its job:
The value of
Xis stored in the Context.The value of
Xis pulled from the Context and passed to the detector block.The output from the detector block is stored in the Context as the features variable.
The value of
Xandfeaturesis pulled from the Context and passed to the encoder block.The output from encoder is put back into the Context as
X, overwriting the old value.The value of
Xis pulled again from the Context and passed to the estimator block.The output from the estimator block is returned.
![digraph G {
subgraph cluster_0 {
label = "pipeline.predict(X)";
b0 [label="detector.produce(X)"];
b1 [label="encoder.produce(X, features=features)"];
b2 [label="estimator.produce(X)"];
b0 -> b1 -> b2 [style=invis];
subgraph cluster_1 {
X1 [label=X group=c];
f1 [label=features group=c];
X2 [label=X group=c];
X1 -> f1 -> X2 [style=invis];
X1 -> X2 [style=dashed];
label = "Context";
}
}
X -> X1;
X1 -> b0 [constraint=false];
b0 -> f1;
{X1 f1} -> b1 [constraint=false];
b1 -> X2;
X2 -> b2 [constraint=false]
b2 -> y
}](../_images/graphviz-74f15df84efdedb1488bfe2b184fbe7cecd8355d.png)
JSON Annotations¶
Like primitives, Pipelines can also be annotated and stored as dicts or JSON files that contain
the different arguments expected by the MLPipeline class, as well as the set hyperparameters
and tunable hyperparameters.
Representing a Pipeline as a dict¶
The dict representation of an Pipeline can be obtained directly from an MLPipeline instance,
by calling its to_dict method.
In [7]: pipeline.to_dict()
Out[7]:
{'primitives': ['sklearn.preprocessing.StandardScaler',
'sklearn.ensemble.RandomForestClassifier'],
'init_params': {'sklearn.preprocessing.StandardScaler': {'with_mean': False},
'sklearn.ensemble.RandomForestClassifier': {'n_estimators': 100}},
'input_names': {},
'output_names': {},
'hyperparameters': {'sklearn.preprocessing.StandardScaler#1': {'with_mean': False,
'with_std': True},
'sklearn.ensemble.RandomForestClassifier#1': {'n_jobs': None,
'verbose': 0,
'warm_start': False,
'class_weight': None,
'n_estimators': 100,
'criterion': 'gini',
'max_features': None,
'max_depth': None,
'min_samples_split': 2,
'min_samples_leaf': 1,
'min_weight_fraction_leaf': 0.0,
'max_leaf_nodes': None,
'min_impurity_decrease': 0.0,
'bootstrap': True,
'oob_score': False}},
'tunable_hyperparameters': {'sklearn.preprocessing.StandardScaler#1': {'with_std': {'type': 'bool',
'default': True}},
'sklearn.ensemble.RandomForestClassifier#1': {'criterion': {'type': 'str',
'default': 'gini',
'values': ['entropy', 'gini']},
'max_features': {'type': 'str',
'default': None,
'values': [None, 'auto', 'log2', 'sqrt']},
'max_depth': {'type': 'int', 'default': None, 'range': [1, 30]},
'min_samples_split': {'type': 'int', 'default': 2, 'range': [2, 100]},
'min_samples_leaf': {'type': 'int', 'default': 1, 'range': [1, 100]},
'min_weight_fraction_leaf': {'type': 'float',
'default': 0.0,
'range': [0.0, 0.5]},
'max_leaf_nodes': {'type': 'int', 'default': None, 'range': [2, 1000]},
'min_impurity_decrease': {'type': 'float',
'default': 0.0,
'range': [0.0, 1000.0]},
'bootstrap': {'type': 'bool', 'default': True},
'oob_score': {'type': 'bool', 'default': False}}},
'outputs': {'default': [{'name': 'y',
'type': 'ndarray',
'variable': 'sklearn.ensemble.RandomForestClassifier#1.y'}]}}
Notice how the dict includes all the arguments that used when we created the MLPipeline,
as well as the hyperparameters that the pipeline is currently using and the complete specification
of the tunable hypeparameters.
If we want to directly store the dict as a JSON we can do so by calling the save method
with the path of the JSON file to create.
In [8]: pipeline.save('pipeline.json')
Loading a Pipeline from a dict¶
Similarly, once the we have a dict specification, we can load the Pipeline directly from it
by calling the MLPipeline.from_dict method.
Bear in mind that the hyperparameter values and tunable ranges will be taken from the dict. This means that if we want to tweak the tunable hyperparameters to adjust it to a specific problem or dataset, we can do that directly on our dict representation.
In [9]: pipeline_dict = {
...: "primitives": [
...: "sklearn.preprocessing.StandardScaler",
...: "sklearn.ensemble.RandomForestClassifier"
...: ],
...: "hyperparameters": {
...: "sklearn.ensemble.RandomForestClassifier#1": {
...: "n_jobs": -1,
...: "n_estimators": 100,
...: "max_depth": 5,
...: }
...: },
...: "tunable_hyperparameters": {
...: "sklearn.ensemble.RandomForestClassifier#1": {
...: "max_depth": {
...: "type": "int",
...: "default": 10,
...: "range": [
...: 1,
...: 30
...: ]
...: }
...: }
...: }
...: }
...:
In [10]: pipeline = MLPipeline.from_dict(pipeline_dict)
In [11]: pipeline.get_hyperparameters()
Out[11]:
{'sklearn.preprocessing.StandardScaler#1': {'with_mean': True,
'with_std': True},
'sklearn.ensemble.RandomForestClassifier#1': {'n_jobs': -1,
'verbose': 0,
'warm_start': False,
'class_weight': None,
'n_estimators': 100,
'criterion': 'gini',
'max_features': None,
'max_depth': 5,
'min_samples_split': 2,
'min_samples_leaf': 1,
'min_weight_fraction_leaf': 0.0,
'max_leaf_nodes': None,
'min_impurity_decrease': 0.0,
'bootstrap': True,
'oob_score': False}}
In [12]: pipeline.get_tunable_hyperparameters()
Out[12]:
{'sklearn.ensemble.RandomForestClassifier#1': {'max_depth': {'type': 'int',
'default': 10,
'range': [1, 30]}}}
Note
Notice how we skipped many items in this last dict representation and only included the parts that we want to be different than the default values. MLBlocks will figure out the rest of the elements directly from the primitive annotations on its own!
Like with the save method, the MLPipeline class offers a convenience load method
that allows loading the pipeline directly from a JSON file:
In [13]: pipeline = MLPipeline.load('pipeline.json')
Intermediate Outputs and Partial Execution¶
Sometimes we might be interested in capturing an intermediate output within a pipeline execution in order to inspect it, for debugging purposes, or to reuse it later on in order to speed up a tuning process where the pipeline needs to be executed multiple times over the same data.
For this, two special arguments have been included in the fit and predict
methods of an MLPipeline:
output_¶
The output_ argument indicates which block within the pipeline we are interested
in taking the output values from. This, implicitly, indicates up to which block the
pipeline needs to be executed within fit and predict before returning.
The output_ argument is optional, and it can either be None, which is the default,
and Integer or a String.
And its format is as follows:
If it is
None(default), thefitmethod will return nothing and thepredictmethod will return the output of the last block in the pipeline.If an integer is given, it is interpreted as the block index, starting on 0, and the whole context after executing the specified block will be returned. In case of
fit, this means that the outputs will be returned after fitting a block and then producing it on the same data.If it is a string, it can be interpreted in three ways:
block name: If the string matches a block name exactly, including its hash and counter number
#nat the end, the whole context will be returned after that block is produced.variable_name: If the string does not match any block name and does not contain any dot character,
'.', it will be considered a variable name. In this case, the indicated variable will be extracted from the context and returned after the last block has been produced.block_name + variable_name: If the complete string does not match a block name but it contains at least one dot,
'.', it will be split in two parts on the last dot. If the first part of the string matches a block name exactly, the second part of the string will be considered a variable name, assuming the format{block_name}.{variable_name}, and the indicated variable will be extracted from the context and returned after the block has been produced. Otherwise, if the extractedblock_namedoes not match a block name exactly, aValueErrorwill be raised.
start_¶
The start_ argument indicates which block within the pipeline we are interested
in starting the computation from when executing fit and predict, allowing us
to skip some of the initial blocks.
The start_ argument is optional, and it can either be None, which is the default,
and Integer or a String.
And its format is as follows:
If it is
None, the execution will start on the first block.If it is an integer, it is interpreted as the block index
If it is a string, it is expected to be the name of the block, including the counter number at the end.
This is specially useful when used in combination with the output_ argument, as it
effectively allows us to both capture intermediate outputs for debugging purposes or
reusing intermediate states of the pipeline to accelerate tuning processes.
An example of this situation, where we want to reuse the output of the first block, could be:
context_0 = pipeline.fit(X_train, y_train, output_=0)
# Afterwards, within the tuning loop
pipeline.fit(start_=1, **context_0)
predictions = pipeline.predict(X_test)
score = compute_score(y_test, predictions)
Pipeline debugging¶
Sometimes we might be interested in debugging a pipeline execution and obtain information
about the time, the memory usage, the inputs and outputs that each step takes. This is possible
by using the argument debug with the method fit and predict. This argument allows us
to retrieve critical information from the pipeline execution:
Time: Elapsed time for the primitive and the given stage (fit or predict).Memory: Amount of memory increase or decrease for the given primitive for that pipeline.Input: The input values that the primitive takes for that specific step.Output: The output produced by the primitive.
If the debug argument is set to True then a dictionary will be returned containing all the
elements listed previously:
result, debug_info = pipeline.fit(X_train, y_train, debug=True)
In case you want to retrieve only some of the elements listed above and skip the rest, you can
pass an str to the debug argument with any combination of the following characters:
i: To include inputs.o: To include outputs.m: To include used memory.t: To include elapsed time.
For example, if we are only interested on capturing the elapsed time and used memory during the
fit process, we can call the method as follows:
result, debug_info = pipeline.fit(X_train, y_train, debug='tm')
Warning
Bear in mind that if we use debug=True or saving the Input and Output,
this will consume extra memory ram as it will create copies of the input data and
the output data for each primitive. For profiling it is recommended using the option
tm as shown in the previous example.